Help
How to use the online tool
The design image guides online tool provides user with the crRNAs/sgRNAs oligo pool targeting non-repetitive region. Here are the simple instructions to run the online tool:
1. Select the cell line you are interested in
2. Enter the gene name you want to image
3. Select the non-repetitive region you are interested in
4. Click “Show Oligo” to see the results.
How to use the results
The results provide users with a pool of spacer sequences. After in vitro transcription into crRNAs or sgRNAs, these sequences enable flexible imaging of non-repetitive genomic regions. For detailed instructions on the Oligo-LiveFISH workflow, please refer to the figure below or the manuscript.
How we design effective guides for chromatin imaging
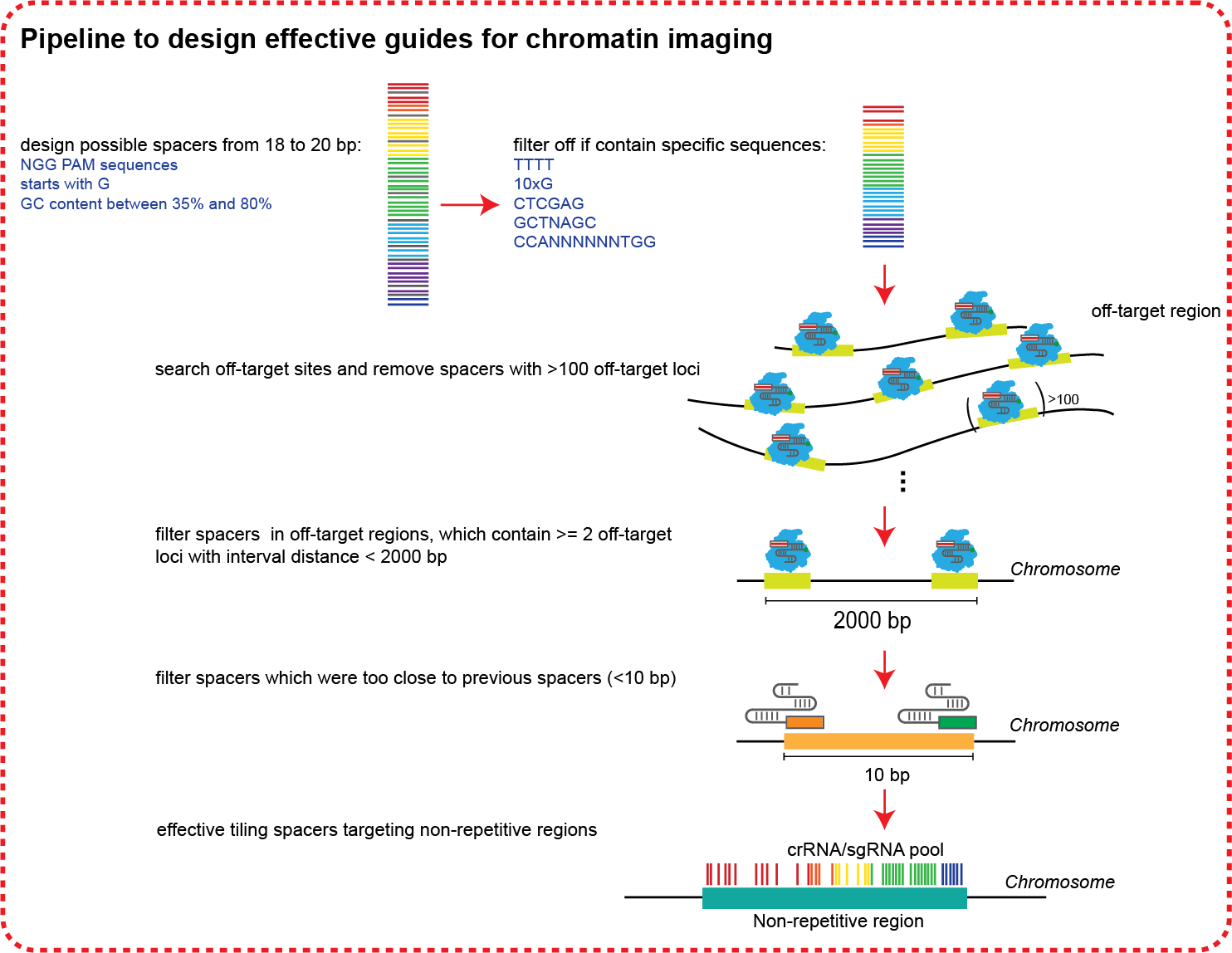
Oligo-LiveFISH Workflow
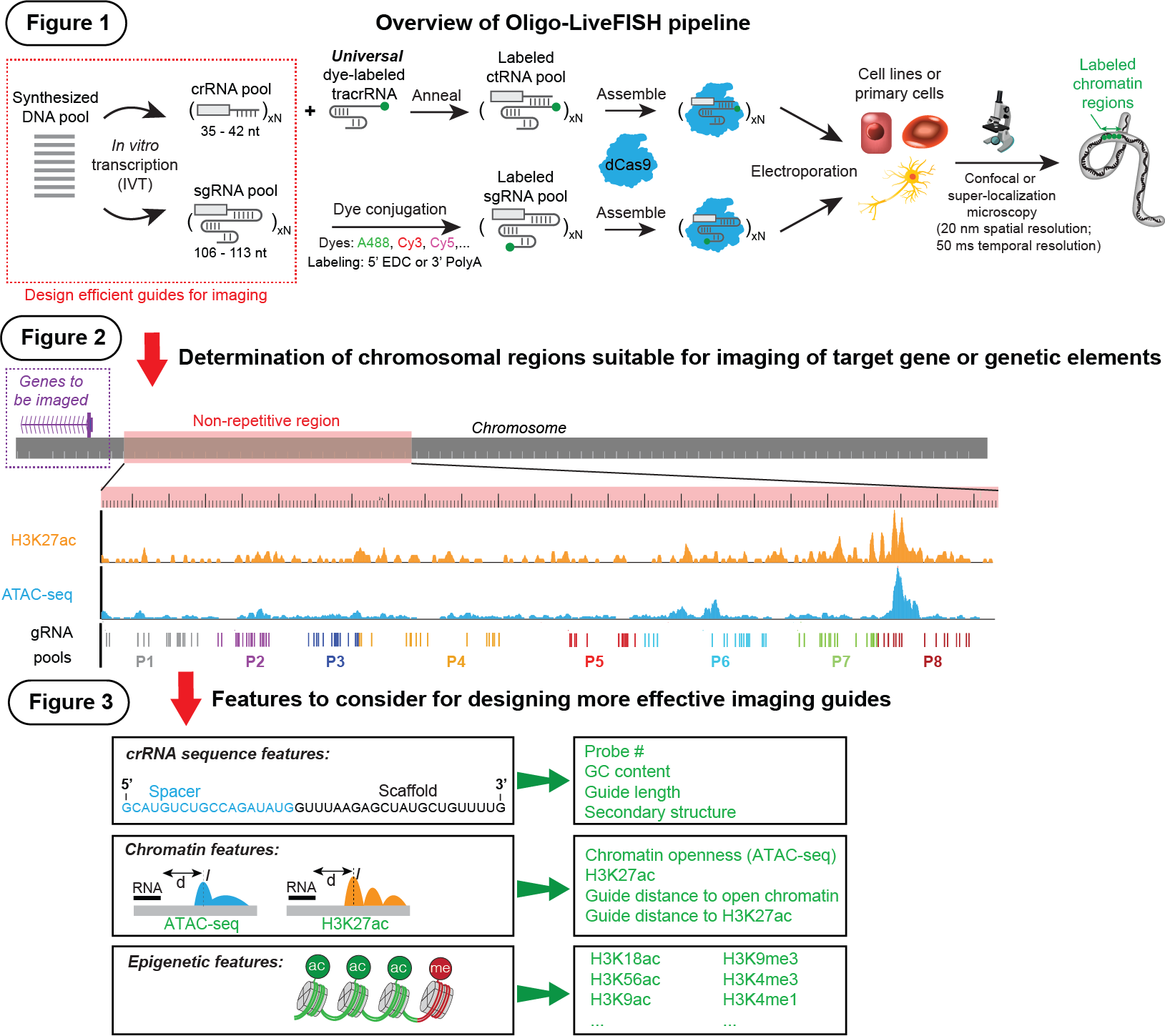
Figure 1: Schematic of Oligo-LiveFISH by delivering a pool of in vitro assembled fRNPs into the live cells for imaging.
Figure 2: Schematic of the position of non-repetitive region near the gene to be imaged. Our pipeline design unique crRNAs/sgRNAs to tile the non-repetitive region.
Figure 3: Features can be used to refine more effective guides for chromatin imaging, including the number of probes, GC content, length and secondary structure of crRNA, and epigenomic profiles such as ATAC-seq intensity, distance to H3K27ac peak,
H3K4me1 intensity and H3K4me3 intensity.